deadwood_categories = [
{'supercategory':'deadwood', 'id':1, 'name': 'Standing'},
{'supercategory':'deadwood', 'id':2, 'name': 'Fallen'},
]
from datetime import date
coco_info = {'description': 'dummydataset for example purposes',
'version': 0.1,
'year': 2022,
'contributor': 'Janne Mäyrä',
'date_created': date.today().strftime("%Y/%m/%d")
}
coco_licenses = {}COCO utilities
COCOProcessor
Utility to transform geospatial data to different COCO formats.
Notes:
- It is possible to specify
min_bbox_areatoshp_to_cocofunction to exclude too small polygons. Default value is 16 pixels - If a detection is a multipart polygon, only the polygon with the largest area is converted to a shapefile.
nor_theta
nor_theta (theta)
Convert angle to simpler format
calc_bearing
calc_bearing (point1, point2)
Get the angle of the rotated bounding box in radians
COCOProcessor
COCOProcessor (data_path:str, outpath:str, coco_info:dict, coco_licenses:list, coco_categories:list)
Handles Transformations from shapefiles to COCO-format and backwards
detectron2_mask_preds_to_coco_anns
detectron2_mask_preds_to_coco_anns (images:list, preds:list)
Process detectron2 prediction to COCO-annotation polygon format. Returns a dict with COCO-style images and annotations
detectron2_bbox_preds_to_coco_anns
detectron2_bbox_preds_to_coco_anns (images:list, preds:list)
Process detectron2 prediction to COCO-annotation polygon format. Returns a dict with COCO-style images and annotations
After tiling the data, COCOProcessor can be used to convert it into a coco style dataset. You need to manually set the coco-info, categories and licenses.
Base version with raw original bounding boxes.
outpath = Path('example_data/tiles/')
coco_proc = COCOProcessor(outpath, outpath, coco_info, coco_licenses, deadwood_categories)
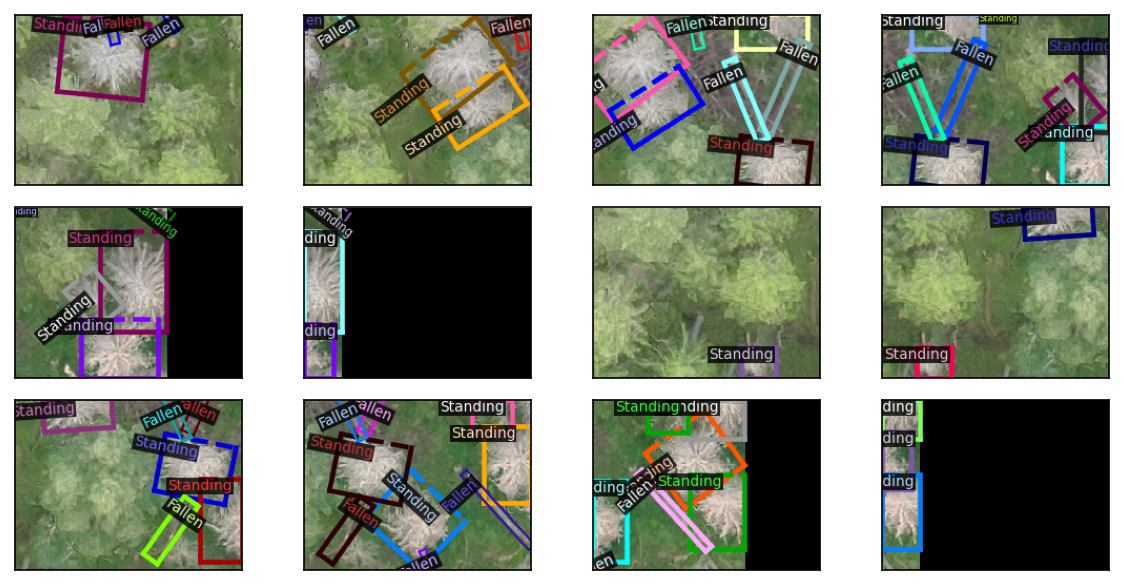
coco_proc.from_shp('label', outfile='coco_norm.json', rotated_bbox=False)You can also do rotated bounding boxes.
outpath = Path('example_data/tiles/')
coco_proc = COCOProcessor(outpath, outpath, coco_info, coco_licenses, deadwood_categories)
coco_proc.from_shp('label', outfile='coco_rot.json', rotated_bbox=True)Use detectron2 to visualize the differences.
from detectron2.data.datasets import register_coco_instances
from detectron2.utils.visualizer import Visualizer
from detectron2.data import MetadataCatalog, DatasetCatalog, build_detection_train_loader, DatasetMapper
import random
import cv2outpath = Path('example_data/tiles/')
register_coco_instances('dummy_norm', {}, outpath/'coco_norm.json', outpath/'images')
register_coco_instances('dummy_rot', {}, outpath/'coco_rot.json', outpath/'images')fig, axs = plt.subplots(3,4, figsize=(8,4), dpi=150)
metadata = MetadataCatalog.get('dummy_norm')
dataset_dicts = DatasetCatalog.get('dummy_norm')
ddicts = dataset_dicts[:12]
for d, ax in zip(ddicts, axs.flatten()):
ax.set_xticks([])
ax.set_yticks([])
img = cv2.imread(d['file_name'])
visualizer = Visualizer(img, metadata=metadata, scale=0.9)
truth = visualizer.draw_dataset_dict(d)
ax.imshow(truth.get_image()[...,::-1])
plt.tight_layout()
plt.show()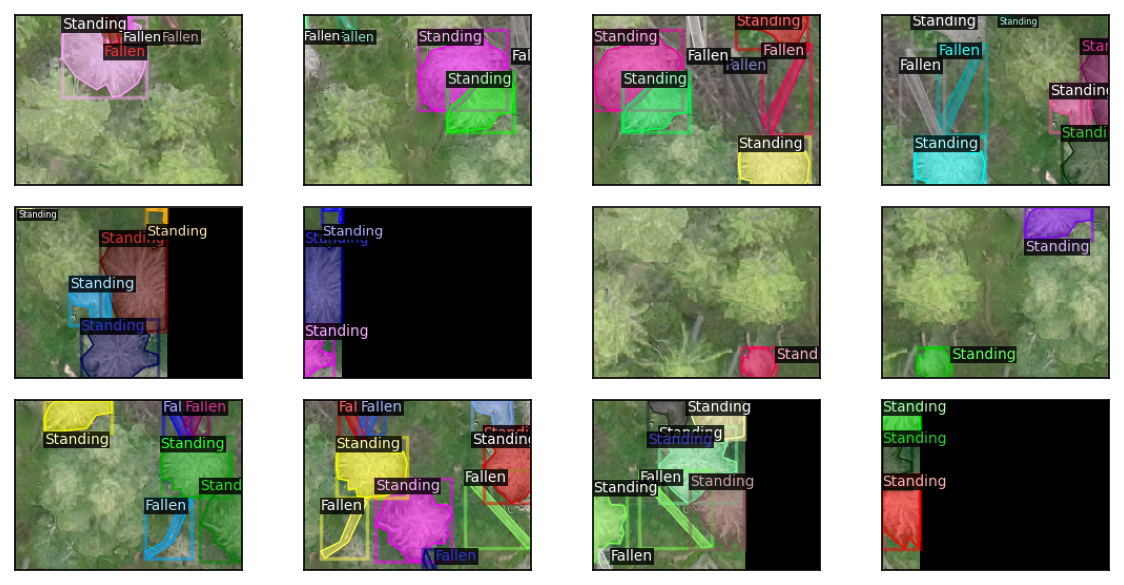
detectron2 doesn’t yet support masks and rotated bounding boxes at the same time.
fig, axs = plt.subplots(3,4, figsize=(8,4), dpi=150)
metadata = MetadataCatalog.get('dummy_rot')
dataset_dicts = DatasetCatalog.get('dummy_rot')
ddicts = dataset_dicts[0:12]
for d, ax in zip(ddicts, axs.flatten()):
ax.set_xticks([])
ax.set_yticks([])
img = cv2.imread(d['file_name'])
visualizer = Visualizer(img, metadata=metadata, scale=0.9)
truth = visualizer.draw_dataset_dict(d)
ax.imshow(truth.get_image()[...,::-1])
plt.tight_layout()
plt.show()